How to Cite
Share
Abstract
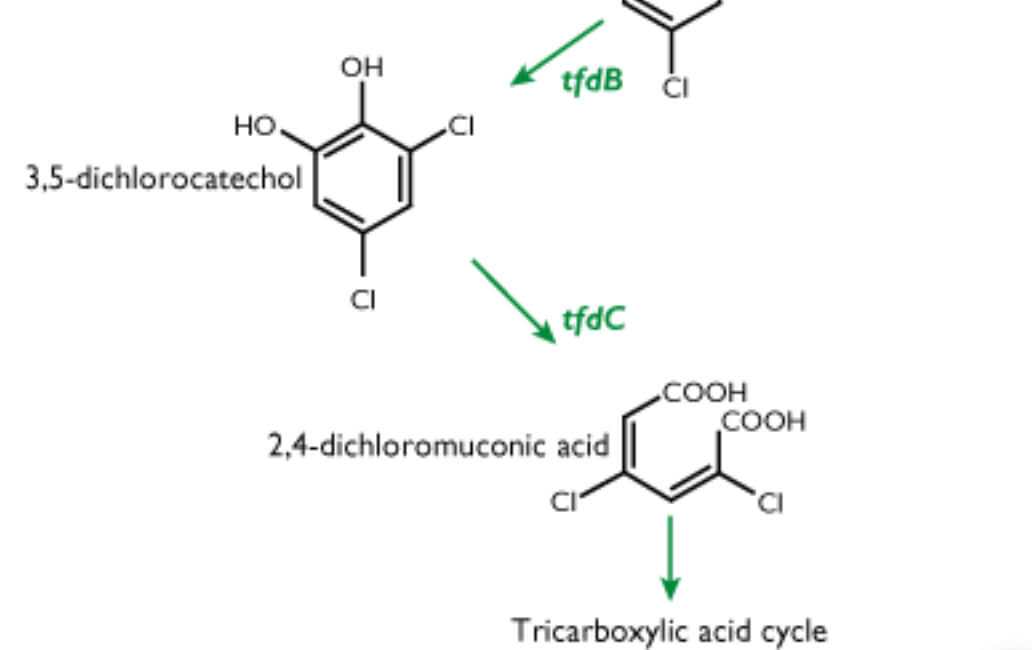
The first DNA-based methods for direct quantification of soil protozoa, and a DNA-based quantification method to describe the spread of phenanthrene-degrading bacteria in soil and freshwater aquifers, have recently been developed at the BIOPRO Research Centre at the Geological Survey of Denmark and Greenland (GEUS). Well-known genes for phenoxyalcanoic acid degradation have been used to monitor the in situ degradation of phenoxyalcanoic acid pesticides. Studies have been initiated on the short-lived mRNA molecules that are expected to provide a shortcut to the understanding of low, yet important, microbial activity in geological samples. This article reviews recent developments in techniques based on analysis of nucleic acids from soils and aquifers. Analytical work has been carried out mainly on soil samples from a former asphalt production plant at Ringe (Fig. 1). The Ringe plant constitutes one of the most polluted industrial sites in Denmark, and is a priority site of studies by the BIOPRO Research Centre. Although rich in carbon, the Ringe subsoil is an oligotrophic environment due to the high content of polycyclic aromatic hydrocarbons (PAH). This is an environment where the supply of nutrients to microorganisms is low, leading to slow growth, low total numbers of microorganisms and small cells. To study microbial communities of oligotrophic environments, analytical methods with low detection limits are needed. Until recently, microorganisms of natural environments were mainly studied by cultivation-dependent methods. However, microorganisms that can be cultured on agar plates are now known to represent only a small fraction of the total microbial community. Modern methods, therefore, need to be based on the detection of biomolecules in the microorganisms rather than being dependent on growth of the microorganisms. The best available techniques are based on DNA and RNA molecules (Fig. 2), which due to their high level of resolution allow closely related organisms or functional genes to be distinguished. In the following review, examples are given of applications of these nucleic acid based methods.
How to Cite
Share
Downloads
Editors: Martin Sønderholm & A.K. Higgins
The Review of Survey activities presents a selection of 23 papers reflecting the wide spectrum of activities of the Geological Survey of Denmark and Greenland, from the microbial to the plate tectonic level.
The Survey's activities in Denmark are documented by ten papers. These include discussion of the [...]










